From advanced chatbots, to AI-generated images, we are seeing applications of generative AI grow by the day. But what actually is generative AI? This article aims to break down the core components of generative AI models and demonstrate applications in the biotech field.
Key concepts: latent factors, self-supervised learning, next token prediction, denoising, fine-tuning
What is Generative AI?
Generative AI is the use of a machine learning model to create new data - this can be new sentences, new images, new molecules (and so on!). There are a variety of machine learning architectures that underpin GenAI models, but in general, they follow two stages: 1) Train a model that represents the patterns in a dataset 2) Sample from this model to create new data points.
Stage 1: Train a model to learn a dataset’s patterns
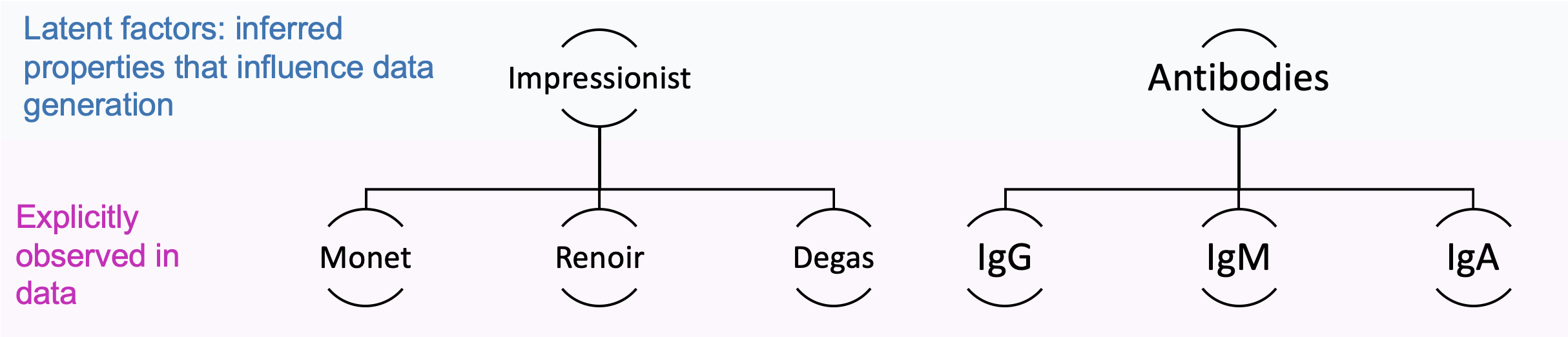
We want a model to identify the underlying properties that influence the data (these are called latent factors) and understand how these properties interact. Latent factors are representations of data that aim to capture higher-level properties. For example, if you asked a model to draw an image in “Monet’s style” you want the model to know that Monet was an impressionist artist, and impressionism correlates with blurry swatches of color - these are properties that define the generation but are not directly labeled from the inputs. An example in biology: if you asked a model to generate a new IgG molecule, you would want the model to know that IgG is a subclass of antibodies - which comes with defined structural patterns.
A model learns these latent factors through a process called self-supervised learning. This training method focuses on taking an input dataset, corrupting it, and learning how to recreate the initial dataset. Corrupting data can look like adding noise to an image, hiding words from a sentence, or masking a protein’s residues. The model iteratively optimizes its parameters until it is able to reliably go from the corrupted data back to the initial dataset. This process of learning how to ‘fill in the gaps’ helps a model learn the generalizable properties of datasets.
Stage 2: Sample from the model to generate new data
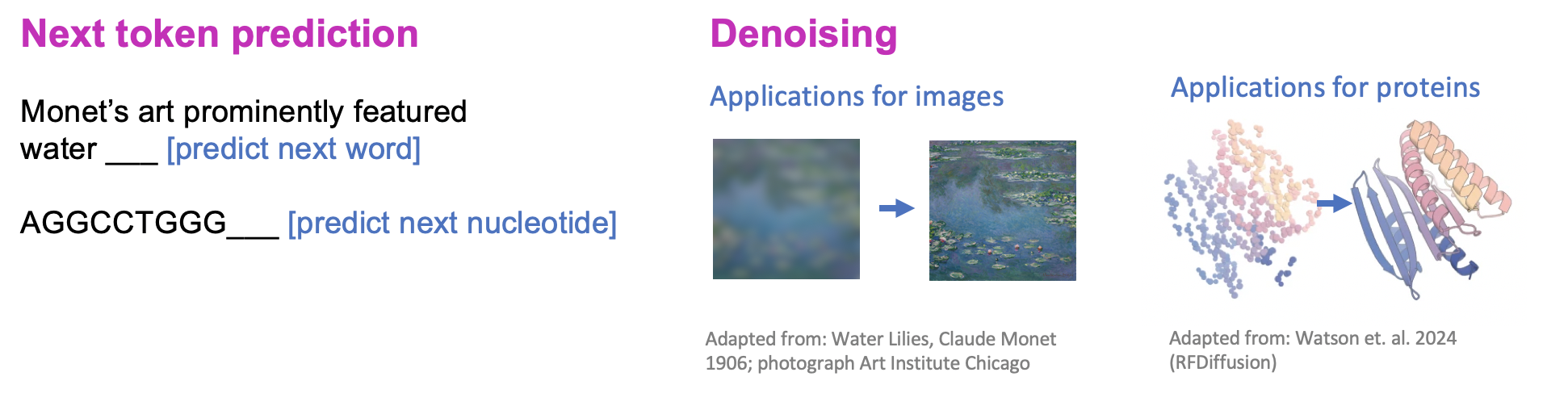
Once we have learned the patterns in our dataset, we can then use that information to generate new data points. One interesting realization of the field is that generating data from scratch is difficult, but generating in small steps is much easier. For text, this can mean reconstructing the next word in a sentence is much easier than predicting the entire next sentence (this process is called next token prediction). For images, it can be easier to first create a blurry outline of the image and then later fill in fine-details, in a process called denoising.
We can also feed a model additional information to guide the type of sample that gets generated. This can look like providing a prompt for an image generator or a question to a chatbot. It can also look like giving the model a specific subclass of data that shares property of what you want to generate, and further training the model on that dataset (fine-tuning). In these cases, the first stage of the training teaches a model general properties of the type of data you want to create, and then the second stage teaches a model specific properties of the subclass you’re interested in. For example, to generate a new antibody, one can start by training a model on a large diverse set of protein sequences to learn general patterns of protein design, and then fine-tuning that model on a smaller set of antibody sequences.
Architectures and Applications
We have skipped over a key part - how exactly do we learn the patterns of latent factors in our data? This depends on the type of data. For text generation, the field has focused on using a specific architecture called a Transformer to model and capture relationships in languages. For image generation, the field has centered on Diffusion models to generate realistic pictures.
Model architectures for biological problems tend to be problem-specific, with scientists typically adapting the methods used in the text and imaging domain. Examples include GenAI models for protein design, small-molecule generation, and cell state representation.
We are at an exciting age in biotech - a time in which we have unprecedented access to large biological datasets, the computational advances in generative AI that allow us to efficiently ingest those datasets, and an ever-present need for faster drug development.
Are there any generative AI x biology applications that you’re particularly excited about? Comment below!