The rise of large foundational models across various domains, including chemical structures, cell types, and proteins, highlights a common challenge: the need for extensive and diverse data. However, these models often spend significant computational resources trying to capture patterns in irrelevant areas of data - for example, trying to model proteins that are not biophysically possible or functional. Biological data is often naturally clustered — for example, proteins form families, and functional molecules are close to existing ones. Because of this, approaches that focus on modeling relevant subsets of the design space can be more efficient. One approach to this, which has been used in large language models like Deepseek, is the Mixture of Experts (MoE) architecture.
Mixture of Experts (MoE): A More Efficient Approach
MoE models improve efficiency by dividing data among specialized expert components. Each expert focuses on specific data patterns, allowing for a refined representation of the relevant subset of data. The expert outputs are then merged back together for downstream tasks, but now with expert-specific knowledge incorporated into the representations. For example, in language models, one expert might learn patterns in punctuation while another focuses on verbs; specializing in these helps the expert better update athe representation of a given word (Zoph et al, 2022).
In addition to efficiency during training, MoE models are particularly more efficient during inference (when the model is used to make predictions), as data only needs to be routed through a relevant subset of parameters rather than the entire model.
How MoE Works
MoE models rely on two key mechanisms:
Gating method - Learns which expert each input should be sent to, often via probability vectors and a learned weight matrix. Inputs can be each sent to a single expert or to a top number of experts (topK routing). The routing can occur at the token level (a portion of a input, for example: a word in a sentence) or at the sample level (the entire input. for example: a protein sequence).
Experts - Specialized neural networks that refine specific subsets of data before merging their outputs. Usually, a researcher will define a type of expert to be used consistently across the model - for example, a common expert is a small feedforward network. While the expert architectures are identical across all the experts used in the model, the parameters are learned separately for each expert, allowing them to specialize in different patterns.
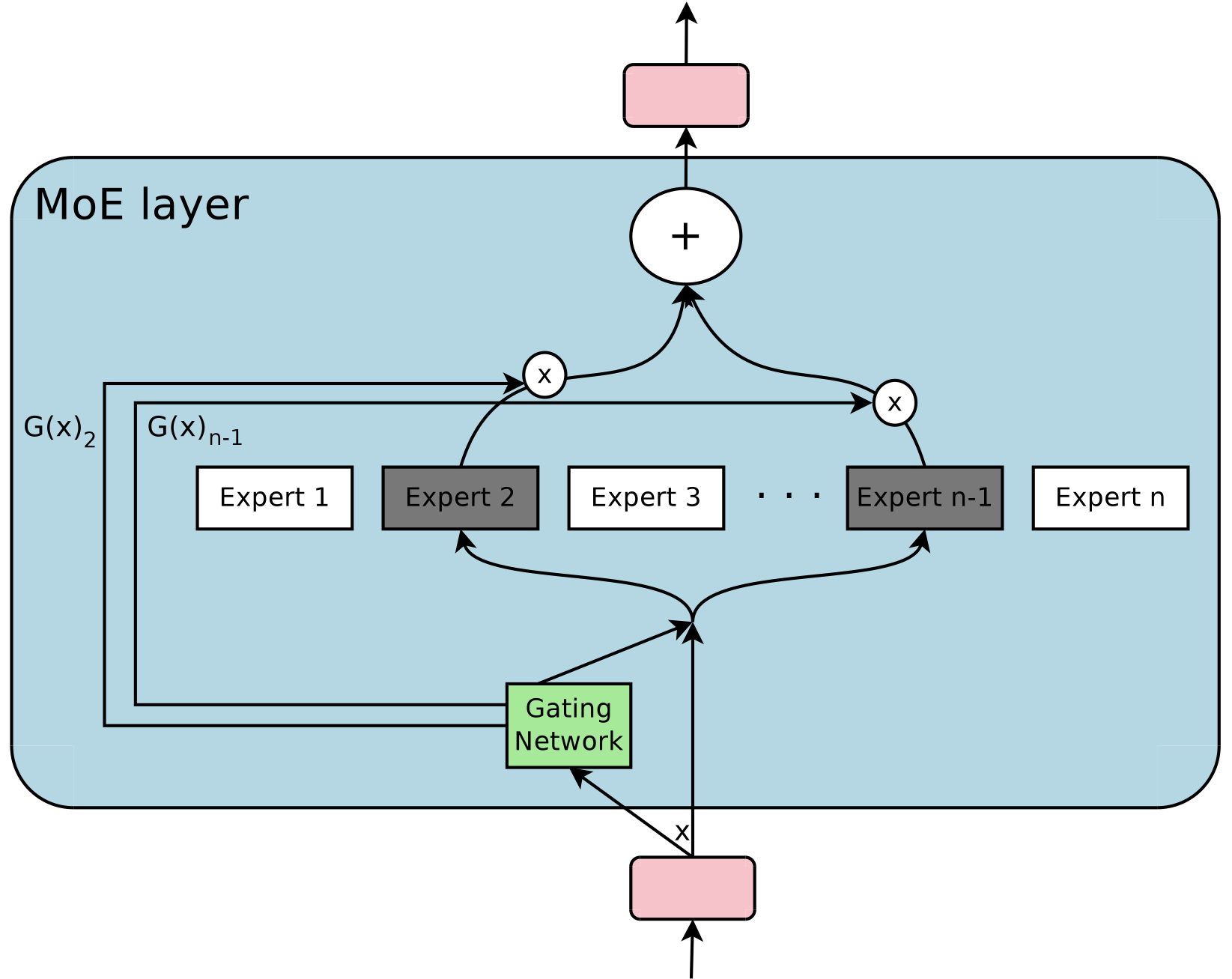 A typical Mixture of Experts architecture. Input data is routed through a gating network that determines which expert(s) should process the data. Each expert specializes in a subset of the data distribution, and their outputs are combined to produce the final result. Image from Shazeer et al, 2017.
A typical Mixture of Experts architecture. Input data is routed through a gating network that determines which expert(s) should process the data. Each expert specializes in a subset of the data distribution, and their outputs are combined to produce the final result. Image from Shazeer et al, 2017.
A key challenge and area of active research for MoEs is load balancing — ensuring inputs are evenly distributed among experts to maximize efficiency. Without constraints, neural networks tend to send all inputs to a one or two large experts. Advances in this field have focused on balancing token distribution across experts to maintain uniformity.
Why it Matters in Biology
MoE models are particularly useful when dealing with clustered latent spaces — common in biological applications. Examples include:
- Modeling proteins, which fall into distinct protein families
- Modeling cell types, which cluster into related classes (Ex: epithelial cells vs neuronal cells)
- Modeling disease subtypes, where patients that have similar disease-related pathways are grouped
In these cases, each expert may focus on a specific class, learn unique patterns within that class, and then merge its findings. For example, each specific protein family might be routed to its own expert. The MoE layers can be further stacked to capture different properties of proteins.
Limitations
MoE models are prone to overfitting, particularly during fine-tuning. Since tokens are divided among multiple experts, each expert processes fewer samples per batch. Similar to how small batch sizes impact the stability of traditional neural networks, MoE models exhibit a higher propensity for overfitting and training instability due to reduced sample diversity within each expert.
Code Implementations
If you’re curious about detailed implementations (how the math actually works under the hood!), check out the code for two examples that I’ve worked on:
- Deep Learning MoE: A minimalist implementation of PyTorch MoE model that I wrote when putting together this article, inspired by DeepSeek’s architecture. I’ve included a notebook that shows how to use the model, train it for a small synthetic dataset, and visualize how it separates data between experts.
- Bayesian MoE: A project from my PhD that applies a mixture of experts model for patient stratification, where gating is defined by a Dirichlet process and experts are Gaussian processes. We released this as an open-source Python package (mogp), and its code can be used as a framework for setting up a Bayesian MoE model.
Conclusion
Mixture of Expert models represent a powerful tool for tackling complex biological problems, offering a way to efficiently model clustered data distributions while maintaining computational efficiency. By intelligently routing data to specialized experts, MoE architectures allow for more precise and scalable solutions in domains like protein modeling, disease classification, and cell type analysis. As advancements continue in balancing, fine-tuning, and optimizing these models, this class of methods can help bridge the gap between biological discovery and computational modeling.
